Ganesh A Viswanathan
Core Faculty
Professor

125, Chemical Engineering
Core Faculty
Professor
IEI Young Engineers Award The Institution of Engineers (India) 2010
IIT Bombay Young Faculty Award 2009
Best Fundamental Paper Award STS-AIChE 2006 & 2007
Office of Science Award Department of Energy 2005
Sigma Xi Best Engineering Graduate Poster Award
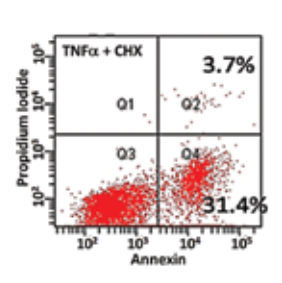
TNFα, a pleiotroiphic cytokine can trigger both pro-apoptotic and prosurvival responses. It has been implicated in cancer treatment, inflammation and autoimmune diseases. The main goal is to perform systems biology mathematical modeling based single-cell experimentation to identify the key molecular signatures of the TNFα signaling network that govern the phenotypic response of a cell exposed to the cytokine. Specifically, the focus is in identifying the optimal set of proteins alteration of whose normal functioning would cause the cell to achieve specific, predictable phenotypic response following exposure to TNFα. TNFα network contains many topological substructures such as interlinked feedback loops which affect the specific phenotype expressed. The lab is interested in understanding the purpose of these substructures and the role it may play in governing the eventual phenotype exhibited by the cell.
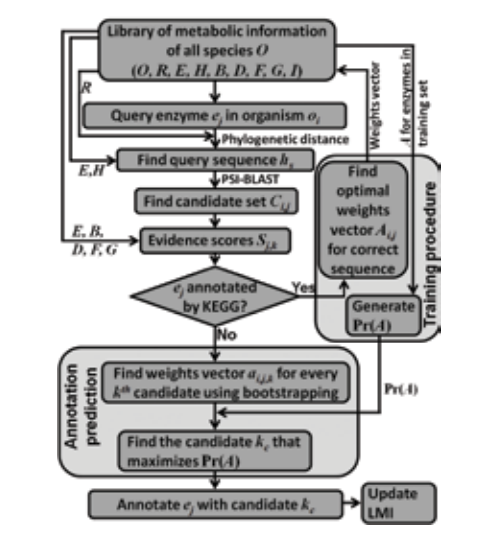
Manual and automated curation approaches are used to develop novel, efficient, systematic methods for construction of signaling, regulatory and metabolic networks. The group is presently involved in developing well-annotated, comprehensive mammalian TNFα signaling network and in identifying the sub-structures and modules that would govern the overall functioning of the network. Recently, the lab has initiated work towards construction and analyses of regulatory and metabolic network of cyanobacteria with a view to develop strategies for engineering the strains for production of biofuel precursors.
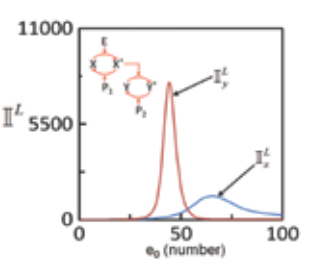
Cells are constantly exposed to inevitable fluctuations or noise from variety of sources. These fluctuations propagate along with the signal and may strongly affect the phenotypic response exhibited by the cells. The lab is interested in characterizing noise propagation in various conserved modules of mammalian signaling networks such as the enzymatic cascades.
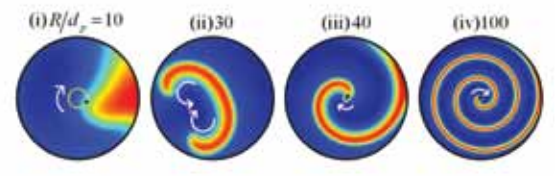
Catalytic packed-bed reactors are workhorse of chemical, petrochemical and pharmaceutical industries. The focus is tp predict the formation of spatiotemporal patterns in non-adiabatic catalytic, packed-bed reactors (PBRs) with an objective to identify the parameter range for safe operation of the reactor. Nonlinear dynamics of the patterns formed in PBRs are characterized using singularity theory and numerical bifurcation analyses.