
Venkatesh Lab
 Understanding Bacterial Metabolism
Understanding Bacterial Metabolism
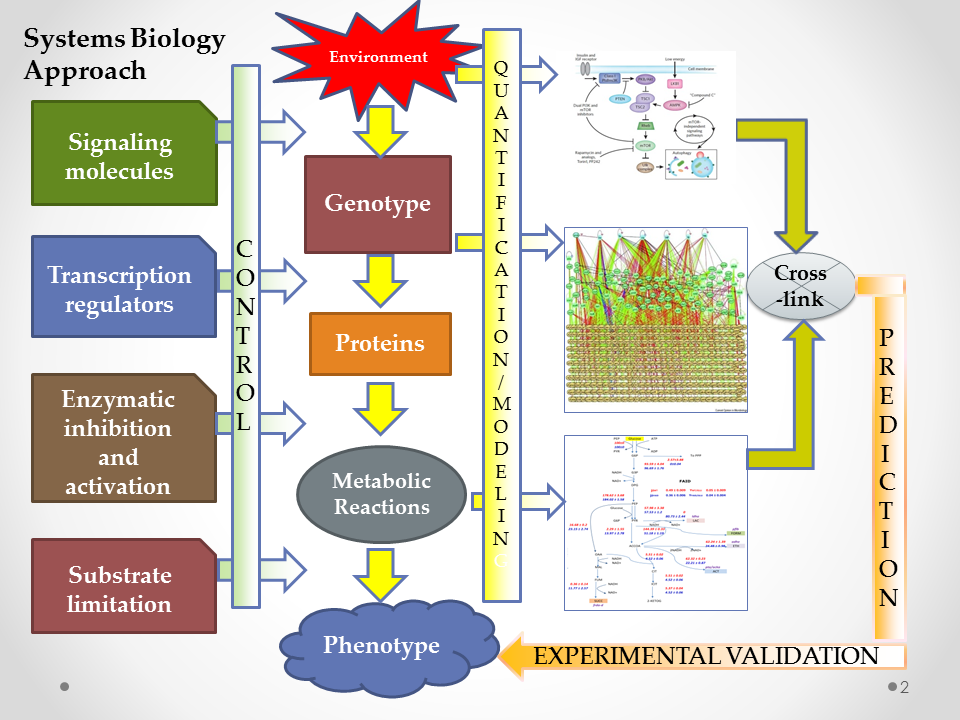
Our group is interested in evaluating the phenotypic characteristics of microbial systems by establishing the fluxes in metabolic networks. We employ flux balance methods and elementary mode analysis to determine the fluxes in the metabolic reaction network. The methodology can further be used to establish the effect of gene deletion and over-expression of enzymes on the fluxes, thereby linking the fluxes to the growth phenotype. These methods have been applied to growth of Escherichia coli, Corynebacterium glutamicum and Saccharomyces cerevisiae. Specifically, we have studied the effect of key transcriptional regulators on the growth of E. coli under different aeration conditions. The study focuses on the relevant question as to whether the global transcriptional regulators have a role in optimizing growth to achieve the three objectives of redox balance, energy generation and biomass synthesis. Further, to establish the connectivity between the genetic networks to the ensuing phenotype, we have developed a steady state modeling formalism for gene expression (SSGES) which includes molecular mechanisms that help in simulating microarray data with high accuracy and can be used to study the effect of both structural and parametric perturbation on gene expression. The expression of TF will then be further connected to the metabolic network.
E. coli metabolism
E. coli is a facultative anaerobic bacterium capable of surviving with (aerobic) or without (anaerobic) oxygen. We study the effect of key transcriptional regulators on the growth of E. coli under different aeration conditions. Transcriptional regulators or factors (TFs) play a key role in regulating the flux distribution in vivo. Many studies enumerate the role of important global transcription factors that regulate metabolism under various conditions.
PUBLICATIONS
Steady state analysis of the genetic regulatory network incorporating underlying molecular mechanisms for anaerobic metabolism in Escherichia coli,
S Srinivasan, KV Venkatesh, Molecular BioSystems, 2014.
Lysine overproducing Corynebacterium glutamicum is characterized by a robust linear combination of two optimal phenotypic states,
M Rajvanshi, K Gayen, KV Venkatesh, Systems and synthetic biology 7 (1-2), 51-62, 2013.
Quantification of metabolism in Saccharomyces cerevisiae under hyper osmotic condition using Elementary mode analysis
Jignesh Parmar, Sharad Bhartiya and K. V. Venkatesh, Journal of Industrial Microbiology and Biotechnology, 2011
Phenotypic Characterization of Corynebacterium glutamicum Using Elementary Modes towards Synthesis of Amino Acids,
Rajvanshi, M., K.V. Venkatesh, , Systems and Synthetic Biology, 2011
Phenotypic characterization of Corynebacterium glutamicum under osmotic stress conditions using elementary mode analysis.
Rajvanshi, M., K.V.Venkatesh, Journal of Industrial Microbiology and Biotechnology , pp. 1-13, 2010.
Quantification of cell size distribution as applied to the growth of Corynebacterium glutamicum
Kalyan Gayen and K V Venkatesh, Microbiological Research, 163, 586-593, 2008.
Elementary mode analysis to study the preculturing effect on the metabolic state of Lactobacillus rhamnosus during growth on mixed substrates,
Kalyan Gayen, Mainsh Gupta and K. V. Venkatesh, In Silico Biology, 7, 0012, 2007.
A phenomenological model to represent the kinetics of growth by Corynebacterium glutamicum for lysine production
Kalyan Gayen and K. V. Venkatesh, Journal of Industrial Microbiology and Biotechnology, 34, 363-372, 2007.
Evaluation of fluxes of elementary modes through linear programming: Applied to Corynebacterium glutamicum
Kalyan Gayen and K.V. Venkatesh, Understanding and exploiting Systems Biology in Biomedicine and Bioprocesses, Editor: Arturo Manjon, 211-222, 2007.
Metabolic flux analysis of biological hydrogen production in Escherichia coli,
Manish S, K.V. Venkatesh and Rangan Banerjee, International Journal of Hydrogen Energy, 32, 3820-3830, 2007.
Steady state approach to model gene regulatory networks- Simulation of microarray experiments,
Subodh Rawool and K V Venkatesh, BioSystems,90(3), 636-655, 2007.
Analysis of optimal phenotypic space using elementary modes as applied to Corynebacterium glutamicum
Kalyan Gayen and K.V. Venkatesh, BMC Bioinformatics, 7,445, 2006.
A Method for the Determination of Flux in Elementary Modes, and its Application to Lactobacillus rhamnosus
M.G. Poolman, K.V. Venkatesh, M.K. Pidcock, and D.A. Fell, Biotechnology and Bioengineering, [88] (5), 601-612, 2004.