
Venkatesh Lab
Whole body Metabolic Models in Humans to Study Metabolic Diseases
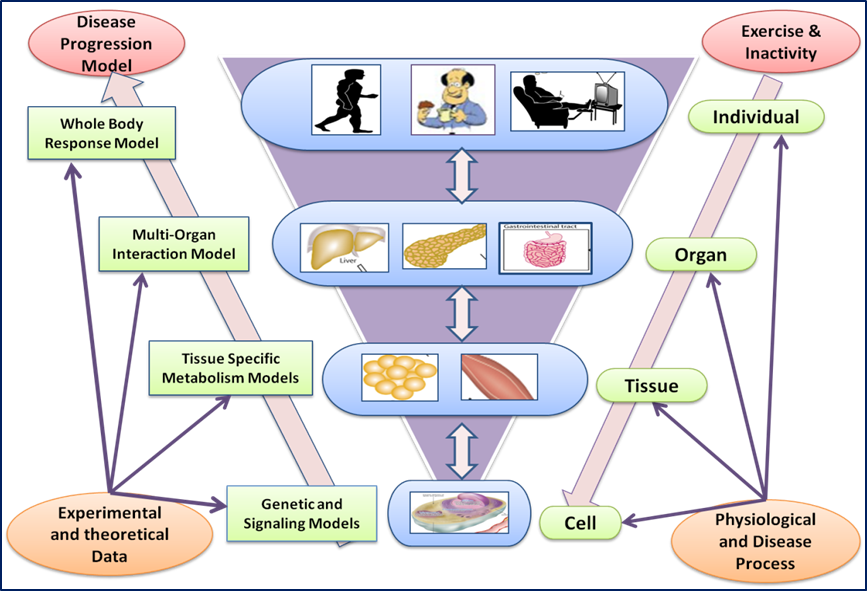 Characterizing disease in mammalian systems is challenging due to the inherent complexity in the system. Diseases such as cancer are found to be multi-factorial and nonlinear. Cancer and Diabetes are among the most complex diseases with multiple complications associated with them. Systems biology techniques enable us to model disease conditions using a holistic approach. The disease condition is an emergent property arising from defects in signaling, genetic and metabolic levels rather than defects in individual biological entities . The main questions we try to answer are –
Characterizing disease in mammalian systems is challenging due to the inherent complexity in the system. Diseases such as cancer are found to be multi-factorial and nonlinear. Cancer and Diabetes are among the most complex diseases with multiple complications associated with them. Systems biology techniques enable us to model disease conditions using a holistic approach. The disease condition is an emergent property arising from defects in signaling, genetic and metabolic levels rather than defects in individual biological entities . The main questions we try to answer are –
(i) what parameter or sets of parameters shift the physiological state of a human from healthy to diseased state?
(ii) How sensitive are these parameters are to perturbations?
(iii) Can the parameters be controlled? If so, how effective is it?
The main challenges in trying to answer these questions lie in trying to – (i) Identify and build a comprehensive hierarchical model of interacting pathways in human metabolic physiology; (ii) Identify disease states and translate existing biological data into quantifiable terms; (iii) Identify sensitive parameters (iv) Analyze the response of the pathway for genetic as well as signal perturbations; (iv) Design metrics to assess performance; (v) Determine steady state as well as dynamic response of the network under parameter perturbations; (vi) Adapt different strategies for analysis such as stochastic or deterministic depending on the nature of the phenomenon under study to name a few.
A whole body metabolic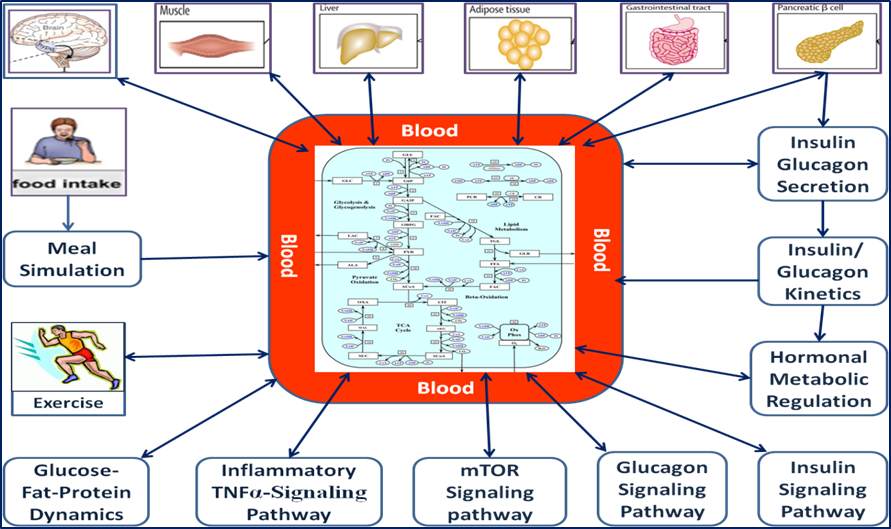 model is under development in our lab comprising of a detailed kinetic models to represent metabolism integrated with signaling pathway with modular analysis towards in silico representation of different organ tissue types such as brain, muscle, liver, adipose tissue, gastro-intestinal tract and pancreatic β cells. The metabolism is connected to blood metabolite concentration and the effect of perturbation in signaling pathway is studied. The components of include models for, (i) Meal Simulation; (ii) Whole body Metabolism; (iii) Insulin-Glucagon signaling Pathway; (iv) Calcium and PKA signaling; (v) Insulin secretion and its Kinetics in Plasma; (vi) Glucose-Fat dynamics in Plasma (Somavanshi, et al.; Kim, et al., 2007; Dall Man, et al., 2007) and integration of these models in a composite form. Thus the model encompasses disease modeling from the level of cells, tissues, and organs to entire body responses using currently available experimental as well as theoretical data. The outputs of signaling pathways were given as the inputs for metabolic pathways. Separate equations were modeled for Glucose transport via Glucose, Fat and Amino acid transporters in various tissues. The kinetic models describing various interactions comprises of around 1000 rate equations including 500 ODEs with 2000 parameters. Parameter values and other constants are collected from the literature wherever available and other parameters are estimated from the optimization of the model under known conditions. The models are then simulated under inactive as well as exercise conditions and sensitivity analysis is performed to discover the key parameters that govern the bio-molecular interactions. Perturbation analysis is performed to get a matrix of parameters for both healthy and disease phenotypes. Such an analysis helps in understanding the system dynamics and aid in identification of potential drug targets.
model is under development in our lab comprising of a detailed kinetic models to represent metabolism integrated with signaling pathway with modular analysis towards in silico representation of different organ tissue types such as brain, muscle, liver, adipose tissue, gastro-intestinal tract and pancreatic β cells. The metabolism is connected to blood metabolite concentration and the effect of perturbation in signaling pathway is studied. The components of include models for, (i) Meal Simulation; (ii) Whole body Metabolism; (iii) Insulin-Glucagon signaling Pathway; (iv) Calcium and PKA signaling; (v) Insulin secretion and its Kinetics in Plasma; (vi) Glucose-Fat dynamics in Plasma (Somavanshi, et al.; Kim, et al., 2007; Dall Man, et al., 2007) and integration of these models in a composite form. Thus the model encompasses disease modeling from the level of cells, tissues, and organs to entire body responses using currently available experimental as well as theoretical data. The outputs of signaling pathways were given as the inputs for metabolic pathways. Separate equations were modeled for Glucose transport via Glucose, Fat and Amino acid transporters in various tissues. The kinetic models describing various interactions comprises of around 1000 rate equations including 500 ODEs with 2000 parameters. Parameter values and other constants are collected from the literature wherever available and other parameters are estimated from the optimization of the model under known conditions. The models are then simulated under inactive as well as exercise conditions and sensitivity analysis is performed to discover the key parameters that govern the bio-molecular interactions. Perturbation analysis is performed to get a matrix of parameters for both healthy and disease phenotypes. Such an analysis helps in understanding the system dynamics and aid in identification of potential drug targets.
PUBLICATIONS
A conceptual review on systems biology in health and disease: From Biological networks to potential therapeutics
Pramod R Somvanshi and K.V. Venkatesh, Perspectives in systems biology, Systems and Synthetic Biology Journal, Springer. Aug 2013.
Control of cholesterol homeostasis by entero- hepatic bile transport–the role of feedback mechanisms
Shekhar Mishra, Pramod R Somvanshi and K.V.Venkatesh, RSC Advances, 2014, 4, 58964